the Seeds of Science
Your source for the latest from the University of Toronto's department of Ecology and Evolutionary Biology
Interview with a software engineer turn biologist
Sitting down with Robert Williamson it is difficult to decide whether he looks more like a software engineer or a biologist. Both, after-all, could be guilty of sporting a t-shirt on which 5 ninjas run on water brandishing Japanese swords, a sixth floundering close by. But he has been both. Born in Alpine, Texas, a small community near Big Bend National Park, Robert began his studies in software engineering at Rose-Hulman Institute of Technology, only to become a biologist. theSeedsofScience put some questions to him about what motivated his change of heart:
What did you want to be when you were 6 years old?
Growing up I had a lot of pets. Our house was a zoo. We had a dozen dogs (not all at once), at least 4 cats, fish, lizards, a pair of turtles, and when I was 8 I got a Quaker parrot (she is my old and grumpy companion still). As you might imagine, I really liked animals, and I wanted to be a veterinarian.
So how did you go from veterinary science to software engineering?
I remember in my 8th grade careers class I took one of those standardized job placement tests; my top three careers were elevator operator, bathroom attendant, and bell hop. Ignoring these suggestions, I continued with my plan to become a vet. But I realized that this would mean I had to help people through emotional times, and see animals that were in a terrible state on a daily basis, and I decided it would be too much for me.
So, a few years later while reading Peter J Bently’s ‘Digital Biology’—a book about how computers are used in biological research—I hit the chapter about evolutionary algorithms. These are programs that construct a population of creatures that face a problem in life, and are played against each other; the most successful get to “mate” to produce the next generation. With each generation, the individuals get better at dealing with the problem. These algorithms are often used to help make efficient timetables and solve difficult engineering problems.
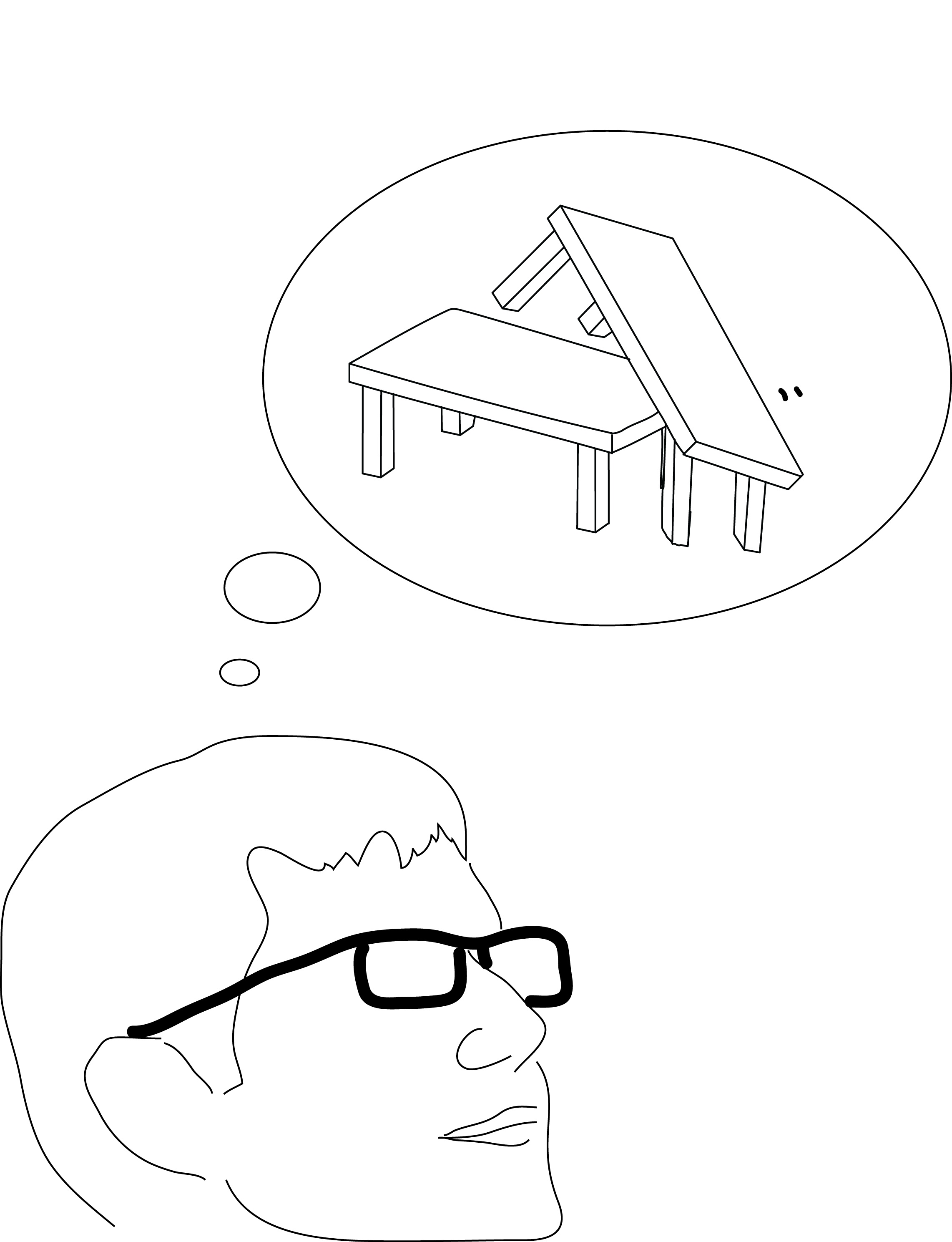
There was this one example where Bently describes an algorithm that evolves a coffee table. The model specifies table height, width, and number of legs; the best “solutions” are those that produce tables that stand up on their own and cannot be easily broken. So Bently let this program run for a while, then took the table with the highest fitness at the end, and had it made. I remember when I saw its picture, I was really wowed by the power of evolution and computing.
So you have a degree in software…what does that mean?
When I first got to Rose-Hulman Institute of Technology I was considering a career in video game design, so I took classes in computer architecture (how does your computer process commands?), programming language concepts and architecture (how to make code pretty, and do cool stuff with it!), database management (how to store lots of data, and retrieve it quickly), and project management (how to make a timetable, schedule, and plan milestones).
What made you switch fields from software to biology?
Over the first couple years at Rose the thought of just being told what projects to work on and coding in a corporate environment started to look less and less appealing. This prompted me to take some summer projects in biology labs, and I realized that breakthroughs in biology are more satisfying to me than breakthroughs in a giant game.
I look at what happens when genes don’t get along. Different genes in your body have different interests, for example you have some genes that are controlled only by the copy you got from your mom and others only from your dad. These genes often have counteracting goals, like “grow fast” and “grow slow”. What you turn out like is determined by how these genes end up resolving their conflict.
How does a background in software help you out in your current work?
I think that there are two things that everyone who wants to do science should know how to do, at least moderately well. The first, statistics, something I still struggle with, and second, programming. Programming can save you endless amounts of time, and help make other people’s programs less like “black boxes”. Programming can help you process data faster, make a quick simulation to test an idea, and, like the code.org video says, it really does “teach you how to think”. Programming is like writing a lab or field protocol: you need to be specific, very clear, and think about scenarios that don’t come up very often.
What do you miss about the life you would have had if you’d stay a software designer?
Well, if I’d stayed in that field I would definitely only work 9-5, and of course I’d be making a boat-load more money.
What of the culture of software design would you bring to the culture of biology?
There have been big pushes in recent years for open-sourced software. This can mean that anybody can submit code, or the code is just available to see, but only a select group works on it. I think that biology can use more of an open-sourced research mind-set. Sometimes we’re so afraid of getting scooped we’re overly cautious. But I feel that things like Haldane’s Sieve encouraging use of arXiv and people like Rosie Redfield who blog about their science in real time are really helping us see what being more open can accomplish.